Very Short Answer Type Questions
Question. Mention the contribution of genetic maps in human genome project.
Answer : Sequencing of genes, DNA finger printing, tracing human history, chromosomal location for disease associated sequences.
Question. Mention the role of the codons AUG and UGA during protein synthesis.
Answer : AUG – codes for methionine / initiation codon.
UGA – termination codon / stop codon.
Short Answer Type Questions
Question. What is aminoacylation ? State its significance.
Answer : Amino acids are activated in the presence of ATP and linked to (cognate) t-RNA.
Carries amino acid to the site of synthesis/reaches amino acids to the respective codon.
Detailed Answer :
Aminoacylation is the process of adding an activated amino acid to the acceptor arm of a transfer RNA.
It is an essential step for the synthesis of protein as it activates the amino acids (amino acid + ATP) and helps in linking them to their cognate tRNA in the presence of an enzyme aminoacyl tRNA synthetase.
Question. State the roles of AUG codon at 5′ end and UAG at 3′ end of a certain m-RNA during translation.
Answer : AUG codon at 5′ end = Start codon (for translation)/codes for methionine.
UAG codon at 3′ end = Stop codon (for translation)/ terminate polypeptide chain.
Detailed Answer :
AUG has dual functions. It codes for Methionine (met) and it also act as initiator codon.
UAG codon functions as stop codon.
Question. One of the salient features of the genetic code is that it is nearly universal from bacteria to humans.
Mention two exceptions to this rule. Why are some codons said to be degenerate ?
Answer : (i) Mitochondrial codons.
(ii) Some protozoans.
Since some amino acids are coded by more than one codon hence it is called as degenerate.
Detailed Answer :
(i) The genetic code is universal except in mitochondria and some protozoans. For example, codon UAA and UGA are termination codons. They do not code for any amino acid but in Paramaecium and a few other ciliates, these codons code for glutamine. Similarly, in yeast mitochondria the codon UGA codes for tryptophan instead of its general terminating character in nuclear genes. The codon AUA codes for isoleucine in nuclear gene but it codes for methionine in mammalian mitochondria.
(ii) When more than one codon code for a single amino acid, it is said to be degenerate codon e.g. UUU and UUC code for amino acid phenyl alanine.
Question. (i) Name the scientist who suggested that the genetic code should be made of a combination of three nucleotides.
(ii) Explain the basis on which he arrived at this conclusion.
Answer : (i) George Gamow.
(ii) There are four bases and 20 amino acids.
(There should be atleast 20 different genetic codes for these 20 amino acids).
Only possible combinations that would meet the requirement in combinations of 3 bases that will give 64 codons.
Detailed Answer :
(i) George Gamow suggested that the genetic code should be made of a combination of three nucleotides.
(ii) There are four types of nitrogenous bases in DNA while the number of amino acids used in protein synthesis is 20. If the code is singlet i.e. consisting of only one nucleotide, this will provide only 4 codons, viz. A, C, G and U/T which are insufficient to code for 20 amino acids. Similarly a combination of two nitrogen bases (doublet codon) will provide codons which are still insufficient for 20 amino acids. Gamow (1954) suggested that the code should be a triplet i.e. made up of different combination of three nucleotides. This will give 4 ×4 × 4 = 64 codons which are more than enough to code 20 amino acids.
Question. Why does the lac operon shut down some time after the addition of lactose in the medium where E.coli was growing? Why low level expression of lac operon is always required?
Answer : After addition of lactose, complete breakdown of lactose to glucose and galactose takes place.
Therefore, there is no more lactose to bind to the repressor protein and the lac operon shuts down.
A very low level of expression of lac operon has to be present in the cell all the time, otherwise lactose cannot enter the cells.
Question. Where is an ‘operator’ located in a prokaryote DNA ? How does an operator regulate gene expression at transcriptional level in a prokaryote ? Explain.
Answer : The operator region is located adjacent to promoter elements / prior to structural gene.
In regulation of gene expression :
switch off – the repressor binds to the operator region and prevents transcription.
switch on – In the presence of inducer the repressor is inactivated (by the interaction with the inducer), operator allows RNA polymerase access to the promoter and transcription proceeds.
Question. Write any three goals of Human Genome Project.
Answer : The three main goals of HGP are :
(i) To determine the sequences of 3 billion base pairs that make up the human DNA.
(ii) To identify all the estimated genes in human DNA.
(iii) To store this information in databases.
Question. Following the collision of two trains a large number of passengers are killed. A majority of them are beyond recognition. Authorities want to hand over the dead to their relatives.
Name a modern scientific method and write the procedure that would help in the identification of kinship.
Answer : DNA fingerprinting is used for identification of kinship.
Procedure :
(i) Variable number of tandem repeats (VNTR’s) are satellite DNA’s that show high degree of polymorphism. They are used as probes in DNA fingerprinting.
(ii) Fragments of DNA from an individual are isolated and cut with restriction endonucleases.
(iii) Fragments are separated according to their size and molecular weight through gel electrophoresis.
(iv) Fragments separated through electrophoresis gel are blotted (immobilised) on a synthetic membrane such as nylon or nitrocellulose.
(v) Immobilised fragments are hybridised with a VNTR probe.
(vi) Hybridised DNA fragments can be detected by autoradiography.
(vii) VNTRs are different in size, ranging from 0.1 to 20 kb. Hence, in the autoradiogram, a band of different sizes will be obtained.
(viii) These bands are the characteristic feature of an individual. They are different in each and every individual except identical twins.
Question.

Given above is the schematic representation of lac operon of E. coli. Explain the functioning of this operon when lactose is provided in the growth medium of the bacteria.
OR
A considerable amount of lactose is added to the growth medium of E.coli. How is the lac operon switched on in the bacteria ? Mention the state of the operon when lactose is digested.
Answer : An operon is a part of genetic material (or DNA), which acts as a single regulated unit of one or more structural genes, an operator gene, a promotor gene, a regulator gene, a repressor and an inducer or compressor.
For Diagram:

In lac operon, when lactose is added, it enters the cell with the help of permease, a small amount of which is already present in the cell. Lactose binds itself to active repressor and changes its structure.
The repressor now fails to bind to the operator.
Then RNA polymerase starts transcription of operon by binding to the promoter site P. All the three enzymes for lactose metabolism are synthesized. Finally all the lactose molecules are used up in the whole process of induction.
It can be understood by the above mentioned figure.
After sometime, when the whole lactose is consumed, there is no inducer present to bind to the repressor. Then the repressor becomes active again, attaches itself to the operator and finally switches off the operon.
Question. Unambiguous, universal and degenerate are some of the terms used for the genetic code. Explain the salient features of each one of them.
Answer : Unambiguous – One codon codes for one amino acid = e.g. AUG (methionine).
Universal – Codon and its corresponding amino acid are the same in all organisms.
Example : Bacteria to human UUU codes for phenylalanine (phe).
Degenerate – Some amino acids are coded by more than one codon.
Example : UUU and UUC code for phenylalanine(phe).
Detailed Answer :
Unambiguous code means that one codon codes for only one amino acid e.g. AUG codes for only methionine.
Universal code means that codon and its corresponding amino acid are the same in all organisms e.g. from bacteria to human, UUU codes for phenylalanine.
Degenerate code means that some amino acids are coded by more than one codon e.g. UUU and UUC code for phenylalanine.
Question. (i) Name the scientist who called tRNA an adapter molecule.
(ii) Draw a clover leaf structure of tRNA showing the following :
(a) tyrosine attached to its amino-acid site
(b) anticodon for this amino acid in its correct site (codon for tyrosine is UCA)
(iii) What does the actual structure of tRNA look like ?
Answer : (i) Francis Crick called t-RNA an adapter molecule.
(ii) For diagram:
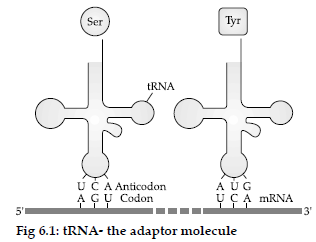
(iii) t-RNA—The actual structure of tRNA looks like a clover leaf having four arms/loops viz: the acceptor arm, ribosomal binding arm, anticodon loop and DHU arm. This clover leaf model of tRNA was proposed by Robert Holley in 1968.
According to Kim (1973), the adapter molecule looks like an L-shaped structure. This is 3-dimensional and is known as the L-shaped model of tRNA.
Question. A criminal blew himself up in a local market when he was chased by cops. His face was beyond recognition. Suggest and describe a modern technique that can help establish his identity.
OR
A number of passengers were severely burnt beyond recognition during a train accident. Name and describe a modern technique that can help establish their identity.
OR
During a fire in an auditorium a large number of assembled guests got burnt beyond recognition.
Suggest and describe a modern technique that can help hand over the dead to their relatives.
Answer : DNA finger printing
Isolation of DNA and digestion of DNA by restriction endonucleases, separation of DNA fragments by gel electrophoresis and transferring (blotting) of separated DNA fragments to synthetic membrane or nitrocellulose or nylon, hybridization using VNTR probe and detection of hybridised DNA fragments by autoradiography, matching the banding pattern so obtained with that of relative.
Detailed Answer :
DNA fingerprinting is the technique of determination of nucleotide sequence of certain areas of DNA, which are unique to each individual.
Step/Procedure in DNA Fingerprinting :
(i) Extraction of DNA — using high speed refrigerated centrifuge.
(ii) Amplification — many copies are made using PCR.
(iii) Restriction Digestion → using restriction enzymes DNA is cut into fragments.
(iv) Separation of DNA fragments → using electrophoresis agarose polymer gel.
(v) Southern Blotting → Separated DNA sequences are transferred to nitrocellulose or nylon membranes.
(vi) Hybridization → The nylon membranes exposed to radioactive probes.
(vii) Autoradiography → The dark bands develop at the probe site.
(viii) Matching the banding pattern so obtained with that of relative.
Question. (a) Explain VNTR and describe its role in DNA fingerprinting.
(b) List any two applications of DNA fingerprinting technique.
Answer : (a) VNTR-
(i) Variable Number of Tandem Repeats.
(ii) used as a probe (because of its high degree of polymorphism).
(b) Forensic science / criminal investigation (any point related to forensic science) / determine population and genetic diversities / paternity testing / maternity testing / study of evolutionary biology.
Long Answer Type Questions
Question. (i) Write any two different levels at which regulation of Gene Expression could be exerted in Eukaryotes.
(ii) Give a labelled schematic representation of ‘‘lac operon’’ in its ‘‘Switched Off’’ position.
Answer : (i) Transcriptional level (formation of primary transcript).
♦ Processing level (regulation of splicing).
♦ Transport of mRNA from nucleus to cytoplasm.
♦ Translational level.
(ii)

Question. (i) How is DNA fingerprinting done ? Name any two types of human samples which can be used for DNA fingerprinting. Explain the process sequentially.
(ii) Mention any two situations when the technique is useful.
Answer : (i) High degree of polymorphism forms the basis of DNA fingerprinting. It involves isolation of DNA, digestion of DNA by restriction endonucleases, separation of DNA fragments by electrophoresis, transferring/blotting of separated DNA fragments to synthetic membranes (nylon or nitrocellulose), Hybridisation using labelled VNTR probes, detection of hybridised DNA fragments by autoradiography.
DNA from blood/hair follicle/skin/bone/saliva/ sperm (Any two).
(ii) Helps as identification tool in forensic applications, in determining population and genetic diversity and helps in paternity testing. (Any two)
Question. (i) Describe the structure and function of a t-RNA molecule. Why is it referred to as an adapter molecule ?
(ii) Explain the process of splicing of hn-RNA in a eukaryotic cell.
Answer : (i) Clover-leaf shaped or inverted L shaped molecules has an anti codon loop with bases complementary to specific codon, has an amino acid acceptor end.
As it reads the code on one hand binds with the specific amino acid on the other hand.
(ii) Introns are removed, exons are joined in a definite order.
OR
Process of splicing shown diagrammatically.
Detailed Answer :
tRNA is a small RNA containing about 80 nucleotides folded over itself in such a way that about 60% of it becomes a double stranded while the rest remains single stranded. This folding of tRNA gives it a clover leaf like structure. tRNA has recognition sites : the anticodon site and the amino acid binding site at 3′ end. Besides this it has two lateral arms, the Ribosomal binding arm and the DHU arm.
(i) Intiator t-RNA recognise start codon (AUG) so it act as adapter molecule that reads the genetic code.
(ii) During the process of splicing of hnRNA, introns are removed (by the spliceosome) and exons spliced (joined) together.

Question. Which methodology is used while sequencing the total DNA from a cell ? Explain it in detail.
Answer : Methodology used : Sequence Annotation :
(i) total DNA from a cell is isolated.
(ii) converted into random fragments of relatively smaller sizes
(iii) and cloned in suitable host using specialized vectors.
(iv) The cloning results in amplification of each piece of DNA fragment.
(v) The fragments are sequenced using automated DNA sequencers,
(vi) these sequences are then arranged based on some overlapping regions present in them.
(vii) This requires generation of overlapping fragments for sequencing.
(viii) Specialized computer based programmes are developed and
(ix) these sequences are subsequently annotated and assigned to each chromosome.
Question. Write the different components of a lac-operon in E. coli., Explain its expression while in an ‘open‘ state.
OR
Explain the role of lactose as an inducer in a lac operon.
Answer : The arrangement where a (Polycistronic) structural gene is regulated by a common promoter and regulatory genes.
Lactose acts as inducer, binds with repressor protein, RNA polymerase freely moves over the structural genes , transcribes lac mRNA , which in turn produce enzymes – transacetylase, permease, ß-galactosidase (by lac z), responsible for digestion of lactose.
// In lieu of above explanation the following diagram can be considered.
For Diagram

Question. (i) Name the scientist who postulated the presence of an adapter molecule that can assist in protein synthesis.
(ii) Describe its structure with the help of a diagram.
Mention its role in protein synthesis.
Answer : (i) Francis Crick
(ii) Clover leaf / inverted L,
Anticodon loop (complementary to codon of mRNA), acceptor end to bind amino acid.
It reads the codons on mRNA with the help of anticodon loop, brings the corresponding amino acid for the formation of polypeptide chain.

Question. Name the major types of RNAs and explain their role in the process of protein synthesis in prokaryote.
Answer : Three types of RNAs :
(i) mRNA (ii) tRNA (iii)rRNA.
Role :
mRNA – Provides the template for protein synthesis by bringing the genetic information from DNA to the site of protein synthesis / ribosome, also provides site to initiate and terminate the process of protein synthesis.
tRNA – Its anti codon loop read the genetic code on mRNA, brings the corresponding amino acid and bound its amino acid binding end on to the mRNA.
rRNA – Forms a structural component of ribosome, (23SRNA) acts as a catalyst / ribozyme for the formation of peptide bond.
Replication
Very Short Answer Type Questions
Question. Name the enzyme involved in the continous replication of DNA strand. Mention polarity of templet strand.
Answer. DNA-dependent DNA polymerase is the main enzyme that using DNA as a templet catalyse polymerisation of deoxynucleotides. Polarity of the templet strand, where replication is continous (i.e., 5′ → 3′ direction) is 3′ → 5′.
Question. Why is it not possible for an alien DNA to become part of chromosome anywhere along its length and replicate normally?
Answer. It is not possible for an alien DNA to become a part of the chromosome anywhere along the length and replicate normally because of absence of origin of replication (ori). It is the sequence where DNA replication starts. This site is also necessary for binding of DNA polymerase to start DNA replication. As this site may not present in all alien DNA molecules hence they cannot replicate normally.
Question. What will happen if DNA replication is not followed by cell division in a eukaryotic cell?
Answer. DNA replication doubles the amount of DNA in a cell and cell division again halves the amount of DNA i.e., maintains the normal amount of DNA in the daughter nuclei. Thus, if DNA replication is not followed by cell division in a eukaryotic cell, then amount of DNA will increase than normal, resulting in abnormal conditions such as polyploidy
Question. Name the enzyme that joins the small fragment of DNA of a lagging strand during DNA replication.
Answer. The enzyme DNA ligase joins the small fragments of DNA of a lagging strand during DNA replication.
Question. Mention the polarity of the DNA strands a – b and c – d shown in the replicating fork given below.

Answer.a – b = 3′ → 5′
c – d = 5′ → 3′
Question. Name the source of energy for the replication of DNA.
Answer. The sources of energy for the replication of DNA are phosphorylated nucleotides or deoxyribonucleoside triphosphates i.e., deATP, deCTP, deGTP and deTTP.
Question. Name the types of synthesis ‘a’ and ‘b’ occuring in the replication fork of DNA as shown below:

Answer.‘a’ – continous synthesis
‘b’ – discontinous synthesis
Question. Name the enzyme and state its property that is responsible for continuous and discontinuous replication of the two strands of a DNA molecule.
Answer. The enzyme DNA-dependent DNA polymerase catalyse the polymerisation of deoxynucleotides.It catalyses polymerisation only in 5′ → 3′ direction thereby resulting in continuous replication on DNA strand with 3′ → 5′ polarity and discontinuous replication on strand with 5′ → 3′ polarity.
Short Answer Type Questions
Question. Discuss the role, the enzyme DNA ligase plays during DNA replication.
Answer. DNA ligase is an enzyme that catalyses the repair of a single strand break by formation of covalent phosphodiester bond between adjacent 3′ hydroxyl and 5′ phosphoryl groups in double stranded DNA. Hence, the enzyme helps in sealing gaps in DNA fragments and acts as molecular glue.
During replication of DNA, short replicated fragments/segments of DNA or Okazaki fragments are present on the lagging strand. These fragments are joined to form a continuous strand with the help of enzyme DNA ligase.
Question. Show DNA replication with the help of a diagram only.
Answer.Diagrammatic representation of DNA replication is as follow:

Question. Draw a neat labelled sketch of a replicating fork of DNA.
Answer. Diagrammatic representation of replicating fork is as follows:

Question. State the dual role of deoxyribonucleoside triphosphates during DNA replication.
Answer. Deoxyribonucleoside triphosphates (or phosphorylated nucleotides) i.e., deATP, deCTP, deGTP and deTTP serve dual purpose during DNA replication.They act as substrates for the replication process as well as provide energy for the polymerisation of nucleotides.
Question. Compare the roles of the enzymes DNA polymerase and DNA ligase in the replication.
Answer.Differences between DNA polymerase and DNA ligase are:
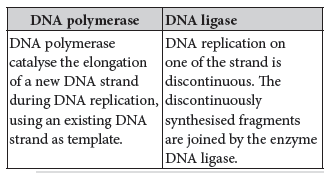
Short Answer Type Questions
Question. Explain the mechanism of DNA replication in bacteria. Why is DNA replication said to be semiconservative?
Answer. DNA replication is a multistep complex process, which in all living cells (including E. coli), requires a dozen enzymes and protein factors. It begins at a particular spot called origin of replication or ori. For DNA replication to proceed, the first requirement is to unwind the double helix, so that the two strands are free to act as templates. Separating the two strands of DNA is accomplished by the helicase enzymes that travel along the helix, opening the double helix by breaking H–bonds between nucleotide pairs as they move. Unwinding also creates a coiling tension in front of the moving replication fork. This tension is reduced by topoisomerases. In prokaryotes topoisomerase is replaced by DNA gyrase.
Prokaryotes have three major types of DNA synthesising enzyme called DNA polymerase I, II and III. All the three DNA polymerases function in 5′ → 3′ direction only for DNA polymerisation and have 3′ → 5′ exonuclease activity.
To initiate DNA synthesis, a small segment of RNA called RNA primer, complementary to the template DNA, is synthesised by a enzyme primase. The two strands of DNA run antiparallel to each other. On one strand the DNA synthesis is continuous in 5′ → 3′ direction, on the other strand, DNA is synthesized in small stretches resulting in discontinuous DNA synthesis. This happens in the opposite direction to the first strand but maintains the overall 5′ → 3′ direction as required. Such a process is also referred to as semi-discontinuous replication. The short stretches of DNA, each primed by RNA are called Okazaki fragments.
RNA primer are then removed, and the gap is filled by DNA synthesis. Both steps are performed by DNA polymerase I. The enzyme ligase then seals these fragments.
The strand which supports the continuous DNA synthesis is the leading strand and the one, which is replicated in short stretches is called the lagging strand.
During DNA replication, one strand of the daughter DNA duplex is derived from the parental strand while the other strand is formed a new.
Since the daughter DNA duplex comprises of one parental and one new strand, thus the replication process is said to be semi-conservative.
Question. Why do you see two different types of replicating strands in the given DNA replication fork?
Explain. Name these strands.

Answer.
The DNA-dependent DNA polymerase catalyse polymerisation in one direction only i.e., 5′ → 3′. Therefore, in the strand with polarity 3′ → 5′ replication is continous whereas on the other strand with polarity 5′ → 3′ replication is discontinous. Therefore, two different types of replicating strands are seen on the DNA replicating fork. The two strands are named leading strand and lagging strand respectively.
Question. (a) Why does DNA replication occur in small replication forks and not in its entire length?
(b) Why is DNA replication continuous and discontinuous in a replication fork?
(c) Explain the importance of ‘orgin of replication’ in a replication fork.
Answer.
(a) For long DNA molecules, since the two strands of DNA cannot be separated in its entire length (due to very high energy requirement), the replication occurs within a small opening of the DNA helix, referred to as replication fork.
(b) DNA-polymerase can polymerise nucleotides only in 5′ → 3′ direction on 3′ → 5′ strand because it adds them at the 3′ end. Since the two strands of DNA run in antiparallel directions, the two templates provide different ends for replication. Replication over the two templates thus proceeds in opposite directions. One strand with polarity 3′ → 5′ forms its complementary strand continuously because 3′ end of the latter is always open for elongation. It is called leading strand. Replication is dicontinuous on the other template with polarity 5′ → 3′ because only a short segment of DNA strand can be built in 5′ → 3′ direction due to exposure of a small stretch of template at one time.
(c) Origin of replication, ‘ori’ is a sequence present in DNA molecule where replication starts. Enzyme helicase acts over the ori site and unwinds the two strands of DNA to start the process of replication.
Question. Answer the following questions based on Meselson and Stahl’s experiment:
(a) Write the name of the chemical substance used as a source of nitrogen in the experiment by them.
(b) Why did the scientists synthesise the light and the heavy DNA molecules in the organism used in the experiment?
(c) How did the scientists make it possible to distinguish the heavy DNA molecule from the light DNA molecule? Explain.
(d) Write the conclusion the scientists arrived at after completing the experiment.
Answer.(a) NH4Cl (Ammonium chloride) was used as a source of nitrogen by Meselson and Stahl in their experiment.
(b) They synthesised light and heavy DNA molecules in the organim (E. coli bacterium) so as to separate different generations on the basis of density gradient centrifugation.
(c) The DNA molecules from each generations, were tested through density gradient centrifugation using cesium chloride. The heavy DNA molecules settled at the bottom whereas successively light DNA molecules settled at the surface. In this way, the DNA molecules were distinguished.
(d) From the experiment scientist concluded that DNA replication is semi conservative in nature.
Question. Describe the experiment that helped demonstrate the semi-conservative mode of DNA replication.
Answer. The work of Mathew Meselson and Franklin Stahl (1958) on E. coli proved semi-conservative replication of DNA.
They first grew bacteria Escherichia coli in a medium containing heavy isotope 15N for several generations. This led to the incorporation of heavy isotope in all nitrogen-containing compounds including bases. They were able to extract the bacterial DNA and centrifuge it in caesium chloride solution. Depending on the mass of the molecule, the DNA would settle out at a particular point in the tube.
The 15N bacteria were then transferred to a growth medium containing the normal, lighter isotope of nitrogen, 14N, where they reproduced by cell division. Meselson and Stahl found that DNA of the first generation was hybrid or intermediate (15N and 14N). It settled in caesium chloride solution at a level higher than the fully labelled DNA of parent bacteria (15N15N). The second generation of bacteria after 40 minutes, contained two types of DNA, 50% light (14N14N) and 50% intermediate (15N14N). At succeeding generation times, the DNA extracts were found to have a lower proportion of 15N as more 14N was incorporated into the bacterial DNA. This observation is possible only if both strands separate during replication and one strand act as template for synthesis of new strand of DNA having 14N. This was conclusive evidence for the semi-conservative method of DNA replication.
Question. (a) Identify the polarity at A and B respectively in the figure given below.

(b) Explain the mechanism the figure represents.
Answer.(a) The polarity of template strands and newly synthesised strands in the given figure can be represented as :

(b) The figure represents the mechanism of DNA replication, during which leading strand and lagging strand are formed on the two different template strands. DNA-polymerase can polymerise nucleotides only in 5′ → 3′ direction. Since the two strands of DNA run in antiparallel directions, the two templates provide different ends for replication. Replication, over the two templates, thus proceeds in opposite directions. On strand with polarity 3′→ 5′ replication is continuous because 3′ end of the latter is always open for elongation. It is called leading strand. Replication is discontinuous on the other template with polarity 5′ → 3′ because only a short segment of DNA strand can be built in 5′ → 3′ direction due to exposure of a small stretch of template at one time. Short segments of replicated DNA are called Okazaki fragments. Okazaki fragments are later joined together by means of enzyme, DNA ligase to form lagging strand.
Question. (a) Why did Meselson and Stahl use 14N and 15N isotopes in the sources of nitrogen present in the culture medium in their experiment?
Explain.
(b) Write conclusion drawn by the them from the experiment.
Answer.(a) Meselson and Stahl used 14N and 15N isotopes for their experiment because 15N is a heavy isotope of nitrogen and can be separated from 14N by density gradient centrifugation using caesium chloride.
Using two different isotopes of nitrogen helped them to isolate the different generations of E. coli bacteria from each other, e.g., they found that DNA of first generation was hybrid (15N and 14N), it settled in density gradient centrifugation at a level higher than the fully labelled DNA of parent bacteria (15N 15N). Succeding generations were also found to settle toward the surface. (b) From the experiment they concluded that DNA replication is semiconservative.
Question. Draw a labelled schematic sketch of replication fork of DNA. Explain the role of the enzymes involved in DNA replication.
Answer.Schematic representation of replication fork is as follows:
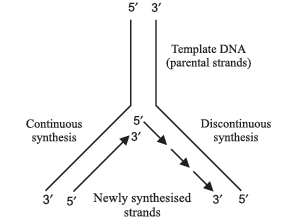
Various enzymes required during DNA replication are helicase, phosphorylase, topoisomerase, DNA polymerase I, II and III, primase and DNA ligase.
Helicase : It acts over the ori site and unzips the two strands of DNA.
Phosphorylase : It is required for phosphorylation of deoxyribonucleotides.
Topoisomerase : It helps in nicking of one strand of DNA to release tension.
DNA polymerase I : It is a major repair enzyme and it has 5′ → 3′ exonuclease activity.
DNA polymerase II : It is a minor repair enzyme.
DNA polymerase III : It does addition and polymerisation of new bases.
Primase : It helps in formation of RNA primer.
DNA ligase : Okazaki fragments are joined by DNA ligase.
Long Answer Type Questions
Question. (a) Draw a labelled diagram of a “replicating fork” showing the polarity. Why does DNA replication occur within such ‘forks’?
(b) Name two enzymes involved in the process of DNA replication, along with their properties.
Answer.(a) Diagrammatic representation of replicating fork is as follows:

Due to high energy requirement whole of DNA does not open in one stretch. The point of separation proceed slowly towards both direction. It gives the appearance of Y-shaped structure called replication fork.
(b) Two enzymes involved in the process of DNA replication are:
(i) Helicase – causes the unwinding of DNA strand.
(ii) Topoisomerase – releases the tension of DNA strand.
Question. Describe Meselson and Stahl’s experiment that was carried in 1958 on E. coli. Write the conclusion they arrived at after the experiment.
Answer.The work of Mathew Meselson and Franklin Stahl (1958) on E.coli proved semi-conservative replication of DNA.
They first grew Escherichia coli bacteria in a medium containing heavy isotope of nitrogen (15N) for several generations. This led to the incorporation of heavy isotope in all nitrogen-containing compounds including bases. They were able to extract the bacterial DNA and centrifuge it in caesium chloride solution. Depending on the mass of the molecule, the DNA would settle out at a particular point in the tube (heavy DNA molecule can be distinguished from normal DNA by centrifugation in cesium chloride density gradient).
The 15N bacteria were then transferred to a growth medium containing the normal, lighter isotope of nitrogen, 14N, where they reproduced by cell division. Extracts of DNA from the first generation offspring were shown to have a lower density, since half the DNA was made up of the original strand containing 15N and the other half was made up of the new strand containing 14N. At succeeding generation times, the DNA extracts were found to have a lower proportion of 15N as more 14N was incorporated into the bacterial DNA. This was conclusive evidence for the semi-conservative method of DNA replication. The conclusion they arrived at after the experiment is that the DNA replication is semiconservative. Semiconservative means that when the double stranded DNA helix was replicated, each of the two double stranded DNA helices of newly synthesised strands consisted of one strand coming from the original helix and one newly synthesised. So, in this way at each replication, one strand of parent DNA is conserved in the daughter while the second is freshly synthesised.
Question. (a) What did Meselson and Stahl observe when
(i) they cultured E. coli in a medium containing 15NH4CI for a few generations and centrifuged the content?
(ii) they transferred one such bacterium to the normal medium of NH4CI and cultured for 2 generations?
(b) What did Meselson and Stahl conclude from this experiment? Explain with the help of diagrams.
(c) Which is the first genetic material? Give reasons in support of your answer.
Answer.(a) (i) Meselson and Stahl observed that in the iiiE. coli, the DNA became completely labelled with N15 when cultured in 15NH4Cl and after centrifugation of this culture they observed that fully labelled DNA of the bacteria settled at the lowest level (near bottom) in the test tube.
(ii) After culturing, one such bacterium, in normal medium of NH4Cl for two generations they observed that its density changed and it showed 50% of light DNA (N14) and 50% intermediate (N15N14).
(b) They concluded that DNA replicates semiconservatively.Diagrammatic representation of semiconservative replication of DNA is as follows:

(c) RNA is the first genetic material as RNAs were the first biocatalysts and essential life processes such as metabolism splicing and translation evolved around RNA.
Question. Explain the process of DNA replication with the help of a replicating fork.
Answer. DNA replication occurs during S-phase of cell cycle. It is a multistep complex process which requires over a dozen enzymes and protein factors. It begins at a particular spot called origin of replication of ori.Replication of DNA is energetically highly expensive.
The main enzyme of DNA replication is DNA dependent DNA polymerase. Deoxyribonucleoside monophosphates occur freely inside the nucleoplasm. They are first phosphorylated and changed to active forms. The phosphorylated nucleotides are deATP (deoxyadenosine triphosphate), deGTP (deoxyguanosine triphosphate), deCTP (deoxycytidine triphosphate) and deTTP (deoxythymidine triphosphate) Enzyme helicase (unwindase) acts over the Ori site and unzips (unwids) the two strands of DNA by destroying hydrogen bonds. Unwinding creates tension in the uncoiled part by forming more supercoils. Tension is released by enzymes topoisomerases. With the help of various enzymes both the strands of DNA become open for replication. However, whole of DNA does not open in one stretch due to very high energy requirement. The point of separation proceeds slowly towards both the directions. In each direction, it gives the appearance of Y-shaped structure called replication fork. RNA primer is a small strand of RNA which is synthesised at the 5′ end of new DNA strand with the help of DNA specific RNA polymerase enzyme called primase.
Prokaryotes have three major types of DNA synthesising enzymes called DNA polymerases III, II and I. In eukaryotes five types of DNA polymerases are found – a, b, g, d and e, but the major three being a, d and e. The two separated DNA strands in the replication fork function as templates. As replication proceeds, new areas of parent DNA duplex unwind and separate so that replication proceeds rapidly from the place of origin towards the other end. RNA primer is removed and the gap filled with complementary nucleotides by means of DNA polymerase I. DNA-polymerase can polymerise nucleotides only in 5′ → 3′ direction on 3′ → 5′ strand because it adds them at the 3′ end. Since the two strands of DNA run in antiparallel directions, the two templates provide different ends for replication. Replication over the two templates thus proceeds in opposite directions. One strand with polarity 3′ → 5′ forms its complementary strand continuously because 3′ end of the latter is always open for elongation. It is called leading strand. Replication is dicontinuous on the other template with polarity 5′ → 3′ because only a short segment of DNA strand can be built in 5′ → 3′ direction due to exposure of a small stretch of template at one time. Short segments of replicated DNA are called Okazaki fragments.
Okazaki fragments are joined together by means of enzyme, DNA ligase. DNA strand built up of Okazaki fragments is called lagging strand. Also refer to answer 70.
Question. (a) Name the stage in cell cycle where DNA replication occur.
(b) Explain the mechanism of DNA replication.Highlight the role of enzymes in the process.
(c) Why is DNA replication said to be semiconservative?
Answer.(a) DNA replication occurs during S-phase of the cell cycle.
(b) DNA replication is a multistep complex process, which requires a dozen enzymes and protein factors. It begins at a particular spot called origin of replication or ori. Separating the two strands of DNA is accomplished by the helicase enzymes that travel along the helix, opening the double helix as they move. Unwinding also creates a coiling tension in front of the moving replication fork, a structure that will be formed when DNA replication begins. This tension is reduced by topoisomerases.
The very important DNA synthesising enzyme is DNA polymerase III. It along with other DNA polymerases (I and II) has the ability to elongate an existing DNA strand but cannot initiate the synthesis.
All the three DNA polymerases function in 5′ → 3′ direction only for DNA polymerisation and have 3 ′ → 5′ exonuclease activity. To initiate DNA synthesis, a small segment of RNA (10 to 60 nucleotides) called an RNA primer complementary to the template DNA is synthesised by a unique RNA polymerase known as primase. While on the one strand the DNA synthesis is continuous in 5′→ 3′ direction, on the other strand, DNA is synthesised in small stretches resulting in discontinuous DNA synthesis. This happens in the opposite direction to the first strand but maintains the overall 5′ → 3′ direction as required. Such a process is also referred to as semi-discontinuous replication. The short stretches of DNA, each primed by RNA are called Okazaki fragments.
RNA primer are then removed, and the gap is filled by DNA synthesis by DNA polymerase I. The enzyme ligase then seals these fragments.
The strand which supports the continuous DNA synthesis is the leading strand and the one, which is replicated in short stretches is called the lagging strand.
(c) DNA replication is said to be semiconservative since daughter DNA duplex comprise of one parental and one newly synthesised strand.