Please refer to Biomolecules Class 11 Biology notes and questions with solutions below. These revision notes and important examination questions have been prepared based on the latest Biology books for Class 11. You can go through the questions and solutions below which will help you to get better marks in your examinations.
Class 11 Biology Biomolecules Notes and Questions
Points to Remember
Biomolecules : All the carbon compounds that we get from living tissues.
Biomicromolecules : Molecules which have molecular weights less than one thousand dalton. They are also known as monomers.
Biomacromolecules : Have molecular weight more than 10000 daltons
(generally 10,000 deltons and above). They are generally polymers.
Biomacromolecules : A biomolecule a with molecular weight in the range of ten thousand daltons and above; found in acid insoluble fraction. e.g. polysaccharides, nucleic acids, proteins and lipids.
Primary and secondary metabolites :
- Primary metabolites have identifiable functions and play important roles in normal physiological process eg. Amino acids, nitrogenous bases, proteins and nucleic acid.
- Secondary metabolites are product of certain metabolic pathways from primary metabolites, eg. carotenoids, drugs, alkaloids, essential oils, rubber, gum, cellulose and resins etc.
Amino acids : Organic compounds containing an amino group and one carboxyl group (acid group) and both these groups are attached to the same carbon atom called a carbon and so they are called oc amino acids.
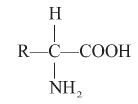
e.g. (1) In Glycine R = H
(2) In alanine R = CH3
(3) In serine R = CH2 – OH
• Twenty types of amino acids.
Amino acid exists in Zwitterionic form at different pHs.

• Based on number of amino and carboxyl groups, amino acids can be:
(i) Aromatic-Tryptophan, phenylalanine and Tyrosine are aromatic (give smell) amino acids.

(ii) Non Polar-Glutamine, tyrosine, serine
Lipids:
Lipids are not strictly macromolecules as their molecular weight do not exceed 800 Da but form a part of the acid insoluble pool.
- Water insoluble, containing C, H, 0.
- Fats on hydrolysis yield fatty acids.
- Fatty acid has a carboxyl group attached to an R group (contains 1 to 19 carbons).
- Fatty Acids : Saturated : With single bonds in carbon chain, e.g., Palmitic acid, butyric acid.
Unsaturated: With one or more double bonds, e.g., oleic acid, linoleic acid. - Glycerol : A simple lipid, is trihydroxy propane.
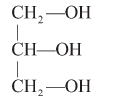
- Some lipid have fatty acids esterified with glycerol.
Example of fatty acid (Palmitic acid) (CH3-(CH2)14—COOH) - They can be monoglycerides, diglycerides and triglycerides.
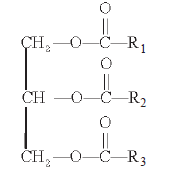
- Triglyceride (R1, R2, R3 are alkyl groups in fatty acids.)
Phospholipids (Lecithin) found in cell membrane and lipids made complex structure in neural tissue. - Phospholipids are compound lipids with phosphorus and a phosphorylated organic compound e.g., Lecithin.

Nucleoside: Nitrogenous base+ Sugar e.g. Adenosine, guanosine.
Nucleotide : Nitrogenous base + Sugar + Phosphate group. e.g. Adenylic acid, Guanylic acid. Thymidylic acid.
Nucleic acids :Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA).
DNAstmcture(Watson and Crick Model): DNAisarighthanded, double helix of two polynucleotide chains, having a major and minor groove. The two chains are antiparallel, and held together by hydrogen bonds (two between A and T and three between C and G). The backbone is formed by sugar-phosphate sugar chain. The nitrogen bases are projected more or less perpendicular to this, backbone and face inside. The pitch is 34A0 . At each step of ascent, the strand turns 36° The rise per base pair is 3.4°A, so one full turn involves ten base pairs.
Protein :proteins are polypeptides.
- They are polymers of aminoacids linked by peptide bond.
- Is a heteropolymer (different monomers repeating ‘n’ number of times).
Structure of Proteins
(a) Primary stmcture: Is found in the form of linear sequence of amino acids.
First amino acid is called N-terminal amino acid and last amino acid is called C-terminal amino acid.
(b) Secondary structm·e :Polypeptide chain undergoes folding or coiling which is stabilized-by hydrogen bonding. Right handed helices are observed; e.g., fibrous protein in hair, nails.
(c) Tertiary stmcture: Long protein chain is folded upon itself like a hollow woollen ball. Gives a 3-dimensional view of protein, e.g., myosin.
(d) Quaternary structure : Two or more polypeptides with their foldings and coilings are arranged with respect to each other, e.g., Human haemoglobin molecule has 4 peptide chains – 2 α and 2 β Subunits.
Monosaccharides are joined by glycosidic bond, right end is reducing and left end is non reducing
Polysaccharides : Are long chain of polymers of monosaccharides.
(a) Star·ch : Store house of energy in plant tissues. Forms helical secondary structures, made of only glucose monomers.
(b) Cellulose : Homopolymer of glucose. It does not certain complex helices.
Cotton fibre is cellulose.
(c) Glycogen :Is a branched homopolymer, found as storage polysaccharide in animals.
(d) Inulin : Is a polymer of fructose.
(e) Chitin: Chemically modified sugar (amino-sugars) N-acetyl galactosamine form exoskeleton of arthropods; heterpolymer.
Metabolic Pathways :
(a) Anabolic pathways : Lead to formation of more complex structure from a simpler structure with the consumption of energy, e.g., Protein from amino acids., also known as biosynthetic pathways.
(b) Catabolic pathway : Lead to formation of simpler structure from a complex structure,
e.g., Glucose → Lactic Acid+ energy
The most important energy currency in living systems is ATP (adenosine tri – phosphate).
“There is no uncatalysed metabolic conversion in living system”
The living state is a non-equilibrium steady state to be able to perform work.
Without metabolism, there cannot be a living state.
Bonds linking monomers in a polymer
Peptide bond- formed between the carboxyl (-COOH) group of one amino acid, and the amino(-NH2) group of the next amino with the elimination of water moiety, (dehydration).
Glycosidic bond- bond formed between two carbon atoms of two adjacent monosaccharides., by dehydration.
Phosphodiester bond- bond formed in nucleic acids where in a phosphate moiety links the 3-carbon of one sugar of one nucleotide to the 5-carbon of the sugar of the succeeding nucleotide. (The bond between phosphate group and hydroxyl group of sugar)
Ezymes : Are biocatalyst.
- Almost all enzymes are proteins.
- Ribozymes-Nucleic acid that behave like enzymes.
- Has primary, secondary and tertiary structure.
- Active site of an enzyme is a crevice or pocket into which substrate fits.
- Enzymes get damaged at high temperatures.
- Enzymes isolated from thermophilic organisms (live under high temperatures) are thermostable.
- Enzymes accelerate the reactions many folds.
- Enzymes lower the activation energy of reactions.
- E + S == ES → EP → E + P
where E = Ezymes, S = Substrate, P = Product

Factors affecting enzyme activity :
(a) Temperature : Show highest activity at optimum temperature. Activity declines above and below the optimum value.
(b) pH : Enzymes function in a narrow range of pH. Highest activity at optimum pH.
(c) Concentration of substrate: The velocity of enzymatic reaction rises with increases in substrate concentration till it reaches maximum velocity (Vmax). Further increase of substrate does not increase the rate of reaction as no free enzyme molecules are available to bind with additional substrate.
KM value : The substrate concentration at which Vmax xis half of a reaction.
Enzyme inhibition : When the binding of a chemical shuts off enzyme activity, the process is called inhibition and chemical is called inhibitor.
Competitive inhibition : Inhibitor closely resembles the substrate in its molecular structure and inhibits the enzyme activity. E.g., inhibition of succinic dehydrogenase by malonate. (Actual is succinic acid).
Classification of enzymes :
- Oxidoreductase/dehydrogenases : Catalyse oxidoreduction between 2 substrates. S reduced+ S’ oxidisedS’ oxidised + S’ reduced.
- Transferases : Catalyse transfer of a group between a pair of substrates.
S – G + S’ → S + S’ – G - Hydrolases : Catalyse hydrolysis of ester, ether, peptide, glycosidic, C-C, P-N bonds.
- Lyases : Catalyse removal of groups from substrates by mechanisms other than hydrolysis. Leave double bonds.
- Isomerases :Catalyse inter-conversion of optical, geometrical or positional tsomers.
- Ligases : Catalyse linking together of 2 compounds.
C-0, C-S, C-N, P-0
Co-factors : Enzymes becomes catalytically become active when it binds to non protein constituent called co-factors. Protein portion of enzyme is called apoenzyme.
- Prosthetic group: These are organic compound which tightly bound to the apoenzyme.
e.g., Haem is prosthetic group in peroxidase and catalase. - Coenzyme : These are organic compounds whose association with the apoenzyme is only transient, usually occurring during the course of catalysis.
e.g., Coenzyme Nicotinamide adenine dinucleotide (NAD) and NADP contain vitamin niacin. - Metal ions : Metal ions form coordination bond with side chains at the active site and at the same time form one or more coordination bond with substrate.
e.g. zinc in enzyme carboxy peptidase.
We hope the above Biomolecules Class 11 Biology are useful for you. If you have any questions then post them in the comments section below. Our teachers will provide you an answer. Also refer to MCQ Questions for Class 11 Biology


